A working molecular script for more recent versions of Blender can be found here: https://github.com/scorpion81/Blender-Molecular-Script/releases
I just tried the most recent version on Windows and I still seeing the ‘core’ error when trying to enable to addon. Has anyone got this to work yet?
Thanks
Works fine for me in Windows 10, Blender 2.92, Molecular 1.1.2 (version from October not 3 days ago).
Thanks for confirming…I’ve not tried it with 2.92, so will give that a go. 
Update: The latest version from 3 days ago, is now working in 2.91 and 2.92
Have tried latest version, and it’s only using one core on my machine. Is this a bug?
Thanks.
He’s currently working as a Houdini artist at Eidos, he probably has no more time to work on it unfortunately 
Works fine here, all CPU threads are used
Let me correct myself: with small particle scale (0.02), very low CPU usage (13%), but with higher scale (1.0), high CPU and RAM usage (100%). Not sure exactly why, but definitely the particle scale value is directly related to the amount of computation required.
Yeah, I don’t understand why.
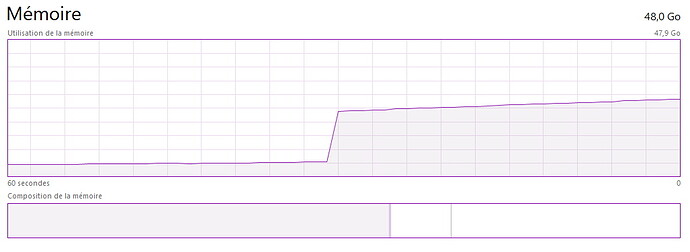
Hey Molecular users, anyone else encounter memory leaks on Windows?
It’s fine until a certain frame in the simulation where the memory gets suddenly overloaded, and continue to rise progressively until full overload.
Also when clearing the simulation cache, the memory is not flushed, it stays full (happens all the time).
Are these known bugs of the addon? Seems to be quite a showstopper.
I’ve encountered these issues with several scenes. On the current one, ~228.000 particles “only”, doesn’t seem to justify such memory overload.
Will test with Linux.
PS: Tested with Linux, Blender crashes a few seconds after starting to bake ![]()
Yeah, have seen the same in windows. Clear cache data and close and open blender.
Do also this before the final caching.
Regards.
Great addon! Very useful. Is there a way to have particles attracted to eachother, as if they were gravitating towards eachother? It seems like there might be some way to do it with the “Linking”, but I just can’t quite figure it out. Thanks!
Raising the number of re-links, I believe, does this.
His site has been down for some time. Look for a newer thread for the location of the update.
Since the blog is down, and I don’t see any support page on github, I decided to post a bug I found trying to activate the add-on in blender 2.93 on Windows 10. Installing the add-on seems to work fine, but when I click the checkbox to activate it I get this error:
Traceback (most recent call last):
File "C:\Program Files\Blender Foundation\Blender 2.93\2.93\scripts\modules\addon_utils.py", line 386, in enable
mod.register()
File "C:\Users\samss\AppData\Roaming\Blender Foundation\Blender\2.93\scripts\addons\molecular\__init__.py", line 42, in register
from . import properties, ui, operators
File "C:\Users\samss\AppData\Roaming\Blender Foundation\Blender\2.93\scripts\addons\molecular\operators.py", line 7, in <module>
from . import simulate, core
ImportError: cannot import name 'core' from 'molecular' (C:\Users\samss\AppData\Roaming\Blender Foundation\Blender\2.93\scripts\addons\molecular\__init__.py)